02/11/22 - Intro to Hypoxia and Shear Stressing
Background:
To gain a better understanding of hypoxia and shear stress before the meeting, we were assigned some videos to watch.
**Endothelial cells = cells that line the walls of the blood vessels
https://www.coursera.org/learn/basic-principles-of-cell-signaling
Video 1: Important Points
- Four types of tissue in the human body: connective, epithelial, muscle, and nervous (our research focuses on epithelial cells)
- Cell Signaling: one cell releases a molecule, which travels to and is received by the target cell. Molecular changes occur within the target cell, and then the structure and function of the target cell can be regulated.
- Examples of signals: hormones, growth factors, nutrients, etc.
- Can use cell signaling to control cell growth, proliferation, differentiation, gene expression, etc.
Video 2: Important Points
- Three major types of cell-cell signaling: electrical, chemical, and gap junctions
- Electrical Signaling: nervous system senses environmental stimuli (like light and touch) and transfers this information through electrical signals that take advantage of membrane potential
- Example: neurons (dendrite –> axon –> axon terminal)
- Chemical Signaling: chemical signals like hormones are produced by specialized cells and released into the bloodstream, which carries them to target cells.
- Useful when two cells are physically separated
- Only target cells have the receptor for the ligand
- Gap Junctions: membrane proteins that allow various molecules to directly pass between adjacent cells
- Shear stress: when blood flows through vessels, it contains little particles, which hit the surface of the endothelial cells, creating a force similar to friction
https://www.youtube.com/watch?v=x8lOO14GFwE
Important Points:
- Hypoxia: when oxygen is not delivered to the tissue
- This is very bad and leads to a lot of serious medical conditions like cyanosis (bluish discoloration of skin), tachycardia (increased heart rate), lethargy, confusion, seizures, coma, and even death
Meeting Notes:
- Clarification about shear stress: shear stress is a good thing. Too much of it can be bad, but that’s more of a long-term issue (like high blood pressure). Too little shear stress is an immediate issue, especially during lung transplants when blood flow stops.
- We discussed how cell signaling pathways work and looked at some examples.
- JAK-STAT signaling pathway
- NF-kB signaling pathway
- HIF signaling pathway
- How to read diagrams:
- A –> B means that A promotes/upregulates B
- A –| B means that A inhibits B
- Talked about log odds: log(test/control)
- Taking the log just makes the data easier to read because it’s centered around 0
- Negative means downregulated while positive means upregulated compared to the control
Homework:
My homework was to read the following paper on hypoxia:
https://pubmed.ncbi.nlm.nih.gov/19101540/#:~:text=Hypoxia%20can%20activate%20the%20endothelium,remodeling%20or%20ischemia%2Dreperfusion%20injury.
02/18/22 - Flamant Paper on Hypoxia
Background:
I read the Flamant paper (linked under last week’s homework) and took notes.
Question: How do inflammatory genes in endothelial cells respond to hypoxia?
Terms to Know:
Leukocyte adhesion: white blood cells sticking to the endothelium when traveling to the site of inflammation or infection
- Cytokines: proteins involved in cell signaling
Results:
- NF-kB target genes (cause inflammation) – expression decreased, less binding of transcription factor to DNA
- HIF-1 target genes (cause inflammation) – expression increased, more leukocyte adhesion to endothelial cells
- Hypoxia is involved in chronic inflammatory diseases
- Since HIF-1 regulates responses to hypoxia, DNA binding activity of HIF-1 increased under hypoxia, and transcription of genes regulated by it also increased
- NF-kB is another major regulator of inflammatory responses but its binding decreased
- Hypoxia did not lead to NF-kB activation in endothelial cells
- In other research, different types of cells did have NF-kB activation during hypoxia, so exact mechanism for NF-kB activation is not clear – may involve ROS (reactive oxygen species)
The paper itself was much more detailed, but these were the most important points.
Meeting Notes:
- We spent a lot of time discussing the papers, what we learned, and any graphs within the papers we didn’t understand
- We took a closer look at the boolean network
- Talked about boolean logic statements
- T && F = F
- T || T || F = T
- T && (T || !F) = T
- Ran some of the code for us
Homework:
Our homework was to use the papers we read and find more relevant papers and try to construct our own boolean network. We were also supposed to download R and R Studio.
02/25/22 - Boolean Networks
Background:
I went through the Flamant paper again but realized it was focusing on umbilical endothelial cells, so I found a different paper that used pulmonary endothelial cells instead. An interesting thing I found was that hypoxia decreased the binding of NF-kB in the umbilical cells but increased binding of NF-kB in pulmonary cells.
This was the additional paper I used: https://www.jimmunol.org/content/177/10/7211
This was the final boolean network I was able to come up with.
Meeting Notes:
- We shared the networks we came up with and talked about how different papers may have different results because of the methods they used
- Specifically, umbilical endothelial cells behave differently than pulmonary endothelial cells (which is what we’re studying), but it’s still okay as long as we notice the things they have in common.
- Also, umbilical endothelial cells are easier to obtain/use, so more studies will use them.
- We took a closer look at the code
- Console vs. Terminal: You can program in R in the console, but the terminal is your computer and won’t understand R
- Parameters vs. Variables: Parameters are usually constant while variables change throughout the code.
- In f(x) = ax + b, x is the variable, a and b are the parameters
- However, we can make a and b variables: f(x, a, b) = ax + b
- For-each loop: goes through each element in a list
Homework:
Try to modify the code to fit my boolean network that I made and read some more literature on endothelial cells and hypoxia.
03/04/22 - Modifying Code
Background:
I found more papers that related to my topic and took notes on information I could add to my boolean network. I have linked the articles and written the information I got from them below.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5979824/#:~:text=As%20the%20SM%2Dlike%20cells,increased%20cell%20proliferation%20and%20migration.&text=Cell%20proliferation%20and%20migration%20following%20hypoxic%20exposure.
- Hypoxia downregulates CD31 & vWF, upregulates a-SMA, Col1A1, Col3A1,
- HIF1a – increases Twist1, a-SMA, Col1A1, decreases CD31
- Its activity increases during pulmonary hypertension
https://www.karger.com/Article/FullText/369709
- Hypoxia increases PDGF-B
- Inhibits apoptosis
- Increases Bcl-2, decreases Bax
- Stimulates phosphorylation of Akt and STAT3
- Inhibitor of PI3K/Akt (LY294002) also decreased STAT3
- PI3K/Akt increases STAT3
https://www.spandidos-publications.com/10.3892/etm.2021.10634
- CXCR4 upregulates PI3K/Akt during hypoxia
https://onlinelibrary.wiley.com/doi/10.1111/jcmm.16386
- MARCH5 increases NO, eNOS, Akt
- Hypoxia decreases MARCH5, decreases eNOS
Updated Boolean Network!
The MARCH5 gene that I added was kind of funny because tomorrow is March 5th 🙂
I also modified Henrique’s code to fit my network. (I’ll try to link it later if I can.)
Meeting Notes:
- Melody and Henrique helped me debug some of my code.
- We also talked about how to get the if statements in the code from the boolean network. Below are some examples.
Homework:
My homework is to read more papers, expand my network, and have something interesting I found in one of my articles that I want to share at the next meeting. Also, I have to make two truth tables with some of the more complicated boolean expressions.
03/11/2022 - Truth Tables
Background:
I read more papers and added to my boolean network.
https://www.cell.com/molecular-therapy-family/nucleic-acids/fulltext/S2162-2531(21)00015-9?_returnURL=https%3A%2F%2Flinkinghub.elsevier.com%2Fretrieve%2Fpii%2FS2162253121000159%3Fshowall%3Dtrue
- The study was about ECFCs (endothelial colony forming cells – regenerate endothelial cells, migrate to areas of injury)
- Hypoxia → miR-130a → VEGFR2, STAT3, HIF1a via inhibition of DDX6 (under hypoxia)
- miR-130 – regulator of pulmonary hypertension
- Increasing miR-130 led to increased angiogenesis
https://www.spandidos-publications.com/10.3892/ijmm.2018.4021
- MLT suppressed the expression of HIF1a and VEGF that was upregulated due to hypoxia
- MLT = melatonin, can inhibit development of cancer, anti-angiogenic
- KC7F2 inhibits HIF1a, ROS, VEGF
- Hypoxia increases ROS, reversed by MLT
- VEGF promoted ROS, which promoted more VEGF (positive feedback) but inconclusive from other studies, inhibited by MLT
- VEGF increases angiogenesis (involved in inflammation, cancer), promotes tumor growth
https://www.sciencedirect.com/science/article/pii/S1347861317301160?via%3Dihub
- Hypoxia activates NF-kB
- TNFa is activated by NF-kB, also increases during hypoxia
Updated Boolean Network:
I also updated the code to include the new connections.
I chose two of the logic statements from my diagram/code to do my truth tables.
Truth Table #1:
Truth Table #2:
Meeting Notes:
- I talked about the papers I had read and the new information I found
- Some of the things in my network, like the LY294002, SB203580, and the KC7F2, were artificially added chemicals that the researchers used in experiments and would not be present naturally, so I will remove them from my diagram and code
- Some of the genes, like HIF-1a, VEGFR, VEGFR2, PI3-k/Akt, and NF-kB are more interesting than the other genes, so I should focus on those
Homework:
My homework was to remove the genes highlighted in red, read more about the genes highlighted in green, and play around with the code to see how removing some genes affects it.
03/18/2022 - Updating Network
Background:
Below is information I found from reading more papers.
https://pubmed.ncbi.nlm.nih.gov/22300081/
HIF-1a increases VEGF (confirmed)
https://link.springer.com/article/10.1007/s10456-020-09748-4
- BMP 2, 4, 6, and 7 are pro-angiogenic
- BMP 9 and 13 are anti-angiogenic
- BMP2/6 → ALK2/3
- VEGF increased BMP2 (confirmed by https://pubmed.ncbi.nlm.nih.gov/12045566/)
- VEGF decreased BMP4
https://www.resmedjournal.com/article/S0954-6111(19)30261-6/fulltext
- Hypoxia → HIF1a → VEGF (confirmed)
- Akt increases VEGF and eNOS
- PDGF increases VEGF
https://www.sciencedirect.com/science/article/abs/pii/S1357272517302558
- NFkB increases ICAM1
- Hypoxia increases eNOS only with HIF
https://www.sciencedirect.com/science/article/pii/S1535610804003022
Hypoxia → HIF1a → VEGF (confirmed again)
Meeting Notes:
- Talked about how MCP-1, IL-1B, and MIP-1B should have an arrow from NF-kB (like TNF-a) because genes need a transcription factor to be expressed (hypoxia cannot directly cause the secretion of proteins)
- The PDGFB upregulating Akt and the VEGF upregulating BMP2 are interesting parts of the network
- I can probably get rid of the microRNA and the DDX6 and just have hypoxia leading directly to HIF-1a
- In intermittent hypoxia, the hypoxia upregulates NOX, which activates HIF-1a, so there would be an extra step there
- However, in continuous hypoxia, the extra step with the NOX is not present
- We looked at sample data sets from Array Express, how to access them, and what they look like
Homework:
My homework is to get rid of anything I think is extraneous and compare my network to Henrique’s to see if there are important genes I’m missing. I can use Kegg to look at some of the pathways. Finally, I can try to get comfortable with looking at data from Array Express.
03/23/2022 - Getting Data
Background:
The following are interesting relationships that I found in Henrique’s network that were not in mine.
- Hypoxia → NOX → NF-kB → HO-1 → CO
- VEGFR2 → Ca (calcium) → NOX
- ALK2 → Smad 1 –| VEGFR2
- TNF → Akt, VEGFR2 → Akt (indirectly)
I read some papers to find out more about these relationships.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3193400/
- Hypoxia → NOX (ROS) → NF-kB confirmed – since ROS is toxic, you have to promote cellular survival, and NF-kB does this (involved in inflammation and immunity)
I tried looking for papers involving calcium but didn’t really find much that talked about the relationship between VEGFR2 and Ca so I’ll probably try to look more next week.
Updated network:
I also tried downloading some files from ArrayExpress.
Meeting Notes:
- We briefly talked about updates on our networks
- Henrique then showed us how to get data from ArrayExpress and put it into a MySQL database
- Since I was on a Windows computer, I had to download WinSCP and PuTTY
- He was telling us what to type to set up the database and troubleshooting all of our errors
Homework:
Our homework is to just read more papers and continue to update our networks.
04/01/2022 - Looking at Databases
Background:
This week, I focused somewhat on the BMPs (bone morphogenic proteins) and related molecules/genes/proteins, etc. Below are the papers I read and what I got out of them.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4149816/
Had a useful chart with lots of signaling pathways, but it was a lot of information and not all of it was necessary. I read through the rest of the paper and noted what I thought was important for my network.
- BMPs promote angiogenesis and mediate shear stress (stress from blood flow) and oxidative stress (accumulation of ROS)
- BMP dysregulation has been linked to pulmonary hypertension
- BMP9 → ALK1 → VEGFR2
- BMP4 → ROS → p38 → apoptosis
- BMP2/4 → eNOS
- BMP2 → ICAM-1 and NF-kB
- Confirmed that ALK2 → Smad 1 –| VEGFR2 (which was also in Henrique’s network)
- Used it to confirm that ROS → p38
- Seemed to have some more useful information, might look at it more in-depth next week
Updated network:
Meeting Notes:
- We discussed the new information we had learned from papers
- ALKs are the receptors for the BMPs, so if a BMP signal is outside the cell, then it binds to the ALK, triggering a signaling pathway inside the cell.
- Sometimes, when there are more BMPs, it can create more receptors for those BMPs
- Henrique had created the databases with the data from ArrayExpress, so we used MySQL to sort through it
- We focused on which genes were significantly upregulated during hypoxia (compared to normoxia)
- We did this using SQL commands like:select symble from SymbleEMTAB3512 where id in (select id from InMatrigelEMTAB3512 where hypoxia_value > 1.5*normoxia_value);
- We were specifically interested in the genes that were transcribed twice as much (or at least had a 50% increase)
- We then used ChEA3 (https://maayanlab.cloud/chea3/) to analyze the gene list we got from the above query.
- The analysis told us which transcription factors were most influential for the genes in our list (it ranked them for us)
- Transcription factors are the proteins that bind to a gene so that it can be transcribed.
- Therefore, if the activity of a gene is high, then it can be inferred that the activity of its transcription factor is also high.
- For example:
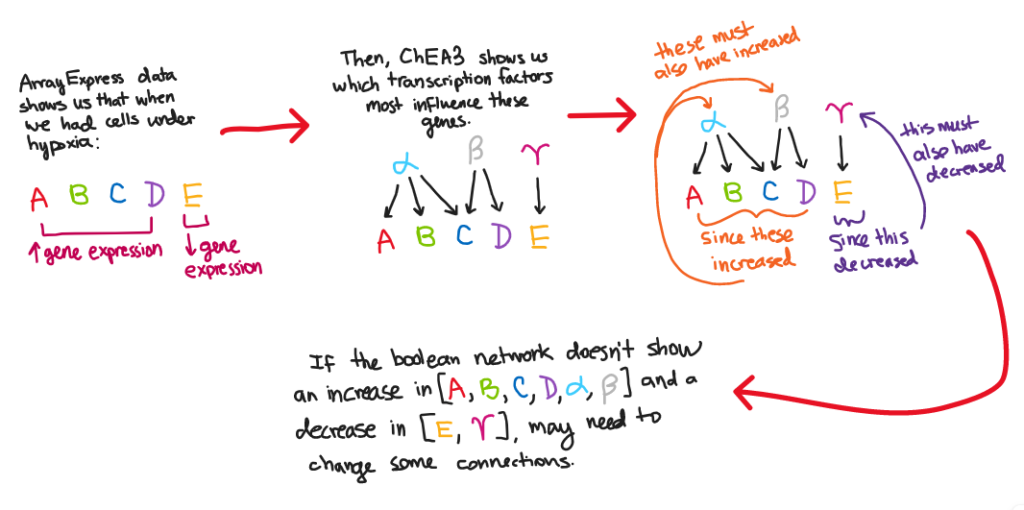
Homework:
04/08/2022 - Writing an Introduction
Background:
Here are the articles I read and the information I got from them:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1140591/\
- ROS necessary for activation of HIF-1a during hypoxia
- p38 → HIF-1a, VEGF
- Hypoxia → IL8 through NFkB
- Hypoxia → p38, PI3k
- p38 –| eNOS
- Lack of eNOS is bad, can cause pulmonary hypertension after exposure to hypoxia
Updated Network:
Meeting Notes:
- We presented what we had learned from our reading this week
- Henrique hadn’t been able to figure out how to get the gene list from the databases so we could put it into ChEA3, so we didn’t work with the data today
- We talked about how to write an introduction to an academic paper, and how it should talk about the purpose of the research
- Whatever we talk about in the introduction (e.g. hypoxia is bad) we have to support by citing a paper to make our claim stronger
Homework:
Our homework is to try to write an introduction to our research, focusing especially on what the purpose is. We should also keep reading more papers.
04/15/2022 - Putting Data Into ChEA3
Background:
This was the introduction I wrote this week:
Hypoxia, which is caused by a lack of adequate oxygen in tissues, has major effects on pulmonary endothelial cell function. It has been shown to play a role in causing pulmonary hypertension and other related diseases, which are among the major causes of death in the world [1]. Hypoxia unavoidably comes into play during lung transplants, because during the time that blood flow is stopped, the pulmonary endothelial cells do not receive oxygen and become hypoxic [2]. Hypoxia can cause endothelial cells to increase surface adhesion molecules and produce reactive oxygen species (ROS) [4], which can be dangerous and cause injury in patients. As a result, hypoxia is one of the major causes of lung transplant failure [2]. It is crucial to determine the pathways that are significantly activated or inhibited in response to hypoxia because it will provide targets in order to minimize the downstream adverse effects of hypoxia. The level of hypoxia-induced gene expression has been correlated with complications after a lung transplantation [2]. One such gene pathway involves the activation of HIF-1a, which led to the proliferation of VEGF, which is known to cause angiogenesis [3]. This study aims to understand these and other key transcription factors, genes, and proteins, such as NF-kB and PI3-k, involved during pulmonary endothelial hypoxia. This can be valuable in increasing survival rate after a lung transplantation.
[1] S.C. Rowan, M.P. Keane, S. Gaine, P. McLoughlin, Hypoxic pulmonary hypertension in chronic lung diseases: novel vasoconstrictor pathways (https://www.sciencedirect.com/science/article/abs/pii/S2213260015005172)
[2] S. Pasnupneti, M.R. Nicolls, Airway hypoxia in lung transplantation (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6688850/)
[3] A.P. Laddha, Y.A. Kulkarni, VEGF and FGF-2: Promising targets for the treatment of respiratory disorders (https://www.resmedjournal.com/article/S0954-6111(19)30261-6/fulltext)
[4] W.A. den Hengst, J.F. Gielis, J.Y. Lin, P.E. Van Schil, L.J. De Windt, A.L. Moens, Lung ischemia-reperfusion injury: a molecular and clinical view on a complex pathophysiological process (https://journals.physiology.org/doi/full/10.1152/ajpheart.00251.2010#:~:text=Physio
logical%20changes%20during%20 ischemia%2d Reperfusion)
Meeting Notes:
- We went over our introductions and talked about maybe focusing on a broader range of topics
- Melody talked about how hypoxia in pulmonary endothelial cells does happen during lung transplants, but I could maybe focus on other applications as well, like clotting
- Henrique showed us the SQL code he used to get the genes where expression during hypoxia >= 1.5*(expression during normoxia) and how he got the actual gene names from the IDs
- He then put the gene list into ChEA3 to look at the Transcription Factors that should also be upregulated and compared it to my model, and he found commonalities (which is good)
Homework:
Our homework is to expand on our introduction and make it longer, and also play around with the SQL queries and data that Henrique and Melody will send us.
04/22/2022 - Revising the Introduction
Background:
This was my expanded introduction:
Pulmonary endothelial cells line the walls of the blood vessels and are essential in regulating blood flow, angiogenesis, the healing of wounds, and inflammatory processes [5]. Hypoxia, which is caused by a lack of adequate oxygen in tissues, has major effects on pulmonary endothelial cell function. Hypoxia can lead to the activation of different signaling cascades within endothelial cells, resulting in the proliferation or inhibition of certain transcription factors, genes, and proteins. For example, hypoxia is known to cause the induction of genes that increase vasoconstriction so that blood is diverted to areas of the lungs with more oxygen [6]. Additionally, upregulated transcription factors like HIF-1a increase levels of the VEGF protein, which is known to cause angiogenesis [3]. As hypoxia plays critical roles in various lung conditions, this study aims to understand and model such pathways within pulmonary endothelial cells that are activated in response to hypoxia.
Hypoxia unavoidably comes into play during lung transplants, because during the time that blood flow is stopped, the pulmonary endothelial cells do not receive oxygen and become hypoxic [2]. Hypoxia can cause endothelial cells to increase surface adhesion molecules and produce reactive oxygen species (ROS) [4], which can be dangerous and cause injury in patients. In fact, the level of hypoxia-induced gene expression has been correlated with complications after a lung transplantation [2]. As a result, hypoxia is one of the major causes of lung transplant failure and is one of the reasons why the five- and ten-year survival rates of lung transplants are relatively low [2]. It is crucial to determine the pathways that are significantly activated or inhibited in response to hypoxia because it will provide targets in order to minimize the downstream adverse effects of hypoxia during a transplant and increase post-transplantation survival rates.
Hypoxia has also been shown to play a role in causing pulmonary hypertension and other related diseases, which are among the major causes of death in the world [1]. In hypoxic conditions, the structure of pulmonary arteries and veins is altered in a process known as pulmonary vascular remodeling [7]. The thickening of the arterial walls increases blood pressure in pulmonary vessels and leads to pulmonary hypertension. It has even been shown that the loss of a single allele of HIF-1a, the transcription factor activated by hypoxia, decreases pulmonary hypertension [8], further connecting the two conditions.
Thrombosis and pulmonary embolism can also be tied back to hypoxia. A pulmonary embolism is when blood clots in a lung artery block the flow of blood to the lung, leading to hypoxia. Often, some parts of the lung end up with much more oxygenated blood than other parts because the clot redistributes the flow of blood [9]. Pulmonary embolisms can thus be life-threatening and cause permanent damage in the lungs, so understanding the mechanisms underlying hypoxia-induced signaling cascades can help minimize deaths due to this condition.
[1] S.C. Rowan, M.P. Keane, S. Gaine, P. McLoughlin, Hypoxic pulmonary hypertension in chronic lung diseases: novel vasoconstrictor pathways (https://www.sciencedirect.com/science/article/abs/pii/S2213260015005172)
[2] S. Pasnupneti, M.R. Nicolls, Airway hypoxia in lung transplantation (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6688850/)
[3] A.P. Laddha, Y.A. Kulkarni, VEGF and FGF-2: Promising targets for the treatment of respiratory disorders (https://www.resmedjournal.com/article/S0954-6111(19)30261-6/fulltext)
[4] W.A. den Hengst, J.F. Gielis, J.Y. Lin, P.E. Van Schil, L.J. De Windt, A.L. Moens, Lung ischemia-reperfusion injury: a molecular and clinical view on a complex pathophysiological process (https://journals.physiology.org/doi/full/10.1152/ajpheart.00251.2010#:~:text=Physiological%20changes%20during%20 ischemia%2d Reperfusion)
[5] J.N. Gonzalez, A.D. Verin, Pulmonary Vascular Endothelial Cells (https://www.intechopen.com/chapters/61198)
[6] K.J. Dunham-Snary, D. Wu, E.A. Sykes, A. Thakrar, L.R.G. Parlow, J.D. Mewburn, J.L. Parlow, S.L. Archer, Hypoxic Pulmonary Vasoconstriction: From Molecular Mechanisms to Medicine (https://pubmed.ncbi.nlm.nih.gov/27645688/#:~:text=Hypoxic%20 pulmonary%20vasoconstriction%20(HPV)%20is,matching%20and%20systemic%20 oxygen%20delivery)
[7] R.M. Tuder, Pulmonary Vascular Remodeling in Pulmonary Hypertension (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5408737/#:~:text=Pulmonary%20vascular%20remodeling%20is%20the,the%20interplay%20of%20inflammatory%20 cells)
[8] M.K. Ball, G.B. Waypa, P.T. Mungai, J.M. Nielsen, L. Czech, V.J. Dudley, L. Beussink, R.W. Dettman, S.K. Berkelhamer, R.H. Steinhorn, S.J. Shah, P.T. Schumacker, Regulation of Hypoxia-induced Pulmonary Hypertension by Vascular Smooth Muscle Hypoxia-Inducible Factor-1α (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3977726/)
[9] S.Z. Goldhaber, C.G. Elliott, Acute Pulmonary Embolism: Part I (https://www.ahajournals.org/doi/10.1161/01.CIR.0000097829.89204.0C)
I also ran the SQL code the Henrique sent us and was able to get the list of gene names, though I was unable to put it into a text file.
Meeting Notes:
- Melody had notes on how to revise my introduction a little bit
- In an actual introduction, it would flow together a little bit more and the paragraphs wouldn’t be about completely separate topics
- Also, the sentence about “This study aims to…” usually goes at the end of the introduction, not at the end of the first paragraph
- One thing that might be useful is to read other papers that used boolean models to see what their reasoning was in choosing boolean models over other types of models, because that might be something we include in the introduction
- Henrique said that after we run the SQL commands in AWS, it won’t let us copy the results of the query into a text file because of security reasons
- An alternative might be that we run the SQL commands but then Henrique sends us the text files with the gene lists so can put it into ChEA3
- Once we get the list of transcription factors from ChEA3, we should compare it to what our boolean network says is activated.
- For example, the following plot was generated after I ran my network in R Studio. It shows that NFkB was initially turned off, but then was turned on and stayed on at about time = 2
Homework:
04/29/2022 - Working on Presentations
Background:
This was the slide I created for my presentation:
I also looked at this paper that also used boolean networks:
https://www.frontiersin.org/articles/10.3389/fphys.2018.01659/full
I added a paragraph to my introduction based on what I read:
When examining such signaling networks, a boolean network was the most useful model. A boolean network is a model in which each node is either turned on or off, and the state of each node depends on the states of the other nodes. The directed edges show how nodes either upregulate or inhibit other nodes. This was the best model to use for this research because it is enough to know whether genes and transcription factors are turned on (significantly upregulated) or turned off (significantly downregulated) in response to hypoxia, and the exact level of expression was less important. In addition, an asynchronous network was used, which means that only one random node is updated at each time step. As a result, there are many possible outcome states and each node continues to affect the other nodes. This is a much more realistic representation of biological systems than a synchronous network would have been.
I also looked at the results from ChEA3. The following table shows the transcription factor that was in my network, its rank according to ChEA3, and the time when it became activated (and stayed activated) in the R Studio simulation:
Meeting Notes:
- We went over what our presentations should look like
- The slide should have fewer words and the images should be bigger, since we’re going to be there to say the words
- Some of the intro information can be shorter just so we have more time to go over our actual model
- For future goals, I can talk about using the model to answer questions, like what pathways cause the most damage to the endothelial cells and what signals can be inhibited to promote cellular survival during hypoxia
- Also, one of the other reasons for using boolean networks is because our data is more qualitative (turned on, turned off) and not really very numerical
Homework:
My homework is to modify my presentation and practice what I’m going to say, and to look into why the nodes in my network turn on and are always turned on (why they never turn off).
05/06/2022 - Practicing Presentations
Background:
This was my final slide for my presentation:
I also figured out my script/what I’m going to say and timed it. It’s about 4 minutes 40 seconds, and we get 5 minutes to speak, so the timing worked out.
Meeting Notes:
- I practiced doing my whole presentation! Melody and Henrique also asked questions at the end just like they will on Tuesday
- Note on how the computer model works: the simulation runs 1000 boolean networks, and each iteration runs for 50 timesteps. The chart shows the average activation of NF-kB over the 1000 boolean networks.
Homework:
My homework is just to make any final changes to my presentation and send it the Melody and Henrique.
05/13/2022 - Next Steps
Background:
The presentation on Tuesday went really well!
Meeting Notes:
- Discussed the presentations
- Scheduling over the summer
- Types of research journals
- Regular journals
- Top-tier journals (Science, Nature, etc.) – not necessarily better science, but more people tend to believe results published in these journals
- Predatory – not scientifically valid
- Explaining the homework (I’ll just do that below)
Homework:
1) I have to look at my model and figure out why everything stays on forever after it’s turned on. One reason was because I had the statement BN[Hypoxia] = BN[Hypoxia], so it was simulating continuous hypoxia. I changed it to BN[Hypoxia] = FALSE so it turns of and then off the rest of the time to simulate intermittent hypoxia. Although this changed some of the graphs, they still stayed on forever, so I have to look at the connections and figure out what’s preventing everything from turning off.
2) Look at the journals I got the information from and make a spreadsheet showing what was manipulated in the experiment (i.e., what the input was) and what the outputs were, also noting what the experiment used (rats, mice, cells, etc.) and organizing it. This could be another way of validating the model.
05/18/2022 - Model Fitting
Background:
I found that the reason HIF1a was always on is because eNOS is always on. My code was:
BN[HIF1a]=(BN[Hypoxia] & !BN[MLT]) | BN[ROS] | BN[p38] | BN[eNOS]
When I turned ROS or p38 off, HIF1a stayed on, which means they were already being turned off. However, when I turned eNOS off, the HIF1a turned off, suggesting that the reason lied with the eNOS. After rereading the paper I found the connection in, I changed the statement to the following:
BN[HIF1a]=(BN[Hypoxia] & !BN[MLT]) | BN[ROS] | BN[p38] | (BN[eNOS] & BN[Hypoxia])
This indicates that eNOS only upregulates HIF1a during hypoxia. This allowed HIF1a to eventually turn off, as shown below:
I also started making a chart of input/output based on the papers I had already read:
Meeting Notes:
- Melody and Henrique pointed out that the reason the eNOS was always on in my network was because one of my statements had a tautology
- My statement for eNOS was:
BN[eNOS]= (BN[March5] | BN[PI3k] | BN[BMP2] | BN[BMP4]) & !BN[p38] - So we looked at my statement for March5:
BN[MARCH5]= !BN[Hypoxia] - And the statement for PI3k:
BN[PI3k]= BN[PDGFB] | (BN[CXCR4] & BN[Hypoxia]) | BN[MARCH5] | BN[Hypoxia] - Because the PI3k statement has March5 | Hypoxia, and because March5 is always the opposite of hypoxia, PI3k is always true (on); therefore, eNOS is always on
- I probably have to see if the statement needs to be modified or if I can just simplify it to TRUE (is it actually always on?)
- My statement for eNOS was:
- For the input/output table, I should just focus on the factors that were directly changed in the experiment (not the intermediate steps in the cascade)
- The purpose is to use the information to make sure that when we run an “experiment” (simulation) on our endothelial cell model and we use the same experiment conditions in a paper (input), we should get the same output they got
- For example, one of the papers had the relationship:
Hypoxia –> PDGFB –> STAT3
Then I should have Hypoxia as the input, and PDGFB and STAT3 both as outputs. PDGFB shouldn’t be listed as an input because it was not artificially added by the experimenters
Homework:
My homework is to keep working on the input/output spreadsheet and examining the logic statements in my code.
05/27/2022 - Combining Networks
Background:
This week, I worked on collecting more input/output data from the papers I already had:
Meeting Notes:
- We compared my network to Isabella’s to see what molecules we had in common
BMP4, P13K/AKT, and eNOS were in common. I had p38 while Isabella had p62, which are both MAP Kinases and are probably related in some way
- We discussed BMPR, BMP2/4, and ALKs. BMPR and the ALKs are probably receptors, but we should look into how they’re connected.
- In my code, ROS was never on, even though it should have been, so we tried to figure out why
Homework:
Our homework is to combine both of our networks into one big network. The network should have a top-down structure, with hypoxia & shear stress leading to receptors, then transcription factors, then genes, and then effects (like proliferation or migration).
06/03/2022 - Combining Networks
Background:
We combined our networks:
These were the new papers we looked at to find more interconnections between hypoxia and shear stress:
https://pubmed.ncbi.nlm.nih.gov/12384983/
- ALK → SMAD
https://pubmed.ncbi.nlm.nih.gov/1618920
- hypoxia → calcium
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4140289/
- GIT1 → eNOS
https://pubmed.ncbi.nlm.nih.gov/32037480/
- Low shear → STAT1
https://www.pnas.org/doi/10.1073/pnas.96.22.12583
- Calcium → eNOS
https://pubmed.ncbi.nlm.nih.gov/20012875/
- Calcium → eNOS (confirmed)
https://www.jbc.org/article/S0021-9258(19)34364-9/fulltext
- HIF1a → ET1
https://www.nature.com/articles/srep10020
- Hypoxia → COX2 → TNFa
https://www.spandidos-publications.com/10.3892/mmr.2020.11060
- Shear stress → PECAM1
https://www.sciencedirect.com/science/article/pii/S0167488918300296
- Shear stress → eNOS
https://www.ahajournals.org/doi/10.1161/01.atv.0000106321.63667.24
Shear stress → ICAM1
Meeting Notes:
- We talked about the new network and potentially using Miro to organize our network better, since all the arrows are getting confusing
- We should also put molecules/genes that have lots of arrows in the middle (like eNOS)
- We also discussed what we should be working on over the next few weeks (I’ll just list that under homework)
Homework:
Our homework is to combine our R code, continue our input/output sheet (separating new papers from papers we used to build the model), starting working on a description of our model (citing all papers used), and finished all connections in the combined model.
06/08/2022 - Using Miro
Background:
We used Miro to organize our model better:
I also finished my input/output data for the papers I used to build my network:
and I also started collecting data for the papers we used to find more connections between the two models:
Meeting Notes:
- We discussed the new model and some possible changes
- All the receptors should be at the top, so we should move VEGFR to the top and just have an arrow pointing up to it
- BMPs bind to BMPR, so the arrow should go from BMP to BMPR
- The BMPs are molecules outside of the cell that then bind to BMPR2, so they should be above everything since they’re extracellular
- Different BMPs have different effects (same with the ALK receptors), so we should separate the ones that do different things
- ERK and p38/p62 are both MAP kinases
- MCP1, IL1B, IL8, and MIP1B are all similar types of signaling molecules (chemokines and cytokines)
Homework:
Our homework is to make the changes to our model, combine our code, continue input/output, and work on the writing portions.
06/17/2022 - Updating the Model
Background:
I separated BMP2 and BMP4 from BMP9 in Miro since they had different effects in the signaling cascade. I also finished my input/output for old papers and added some new papers.
Updated Miro network:
Updated input/output for old papers:
Updated input/output for new papers:
Meeting Notes:
- Double check the connection in the network with SMAD –> BMP. Might go the other way
- Combine BMPR and the ALKs because they are all receptors for the BMP molecules
Homework:
Our homework is to combine our code and write the written description of the model.
06/24/2022 - Combining Code
Background:
Isabella and I combined our R code and updated all of the new connections and molecules. We also worked on our model descriptions.
Here is my description for my side of the model (hypoxia):
Hypoxia activates HIF1a, or hypoxia-inducible factor 1-alpha, which is a transcription factor that plays a large role in endothelial cell response to hypoxia [1]. HIF1a is known to activate several target genes, including PDGFB [2]. PDGFB is also activated by the transcription factor Twist1 [3]. Interestingly, HIF1a has been shown to upregulate Twist1 [4]. The PDGFB gene encodes a protein that binds to the PDGFR receptor and is involved in cell proliferation and differentiation [5]. PDGFB also promotes the expression of the genes VEGF [6], BCL2 [7], and PI3k/Akt [7]. VEGF, or vascular endothelial growth factor, is an important regulator of angiogenesis that binds to the receptor VEGFR2 [8] and promotes the production of reactive oxygen species (ROS) in a positive feedback loop where VEGF-induced ROS production further increases VEGF production [9]. ROS is also known to increase the expression of the kinases p38 and p62 []. BCL2 encodes a family of proteins that primarily serve as regulators of apoptosis [10]. The PI3k/Akt pathway promotes the production of VEGF [6], eNOS [6], and the transcription factors STAT1 and STAT3 [7], and is a regulator of cell proliferation and survival. eNOS is an enzyme responsible for the majority of NO production in endothelial cells and is critical for maintaining homeostasis [11]. STAT1 and STAT3 are essential for angiogenesis as a response to hypoxia, especially because they activate VEGF [12]. HIF1a is also known to directly promote VEGF [1], as well as the gene CXCL8, which produces the chemokine IL-8 [13]. IL-8 is a pro-inflammatory protein that recruits immune cells to the site of inflammation [14]. a-SMA, which increases mechanical tension in endothelial cells [15], and the genes Col1A1/Col3A1, which provide instructions for the production of collagen, are both also upregulated by HIF1a [4]. HIF1a inhibits the production of CD31 [4], a protein responsible for leukocyte adhesion to the endothelium during inflammation, which is encoded by the PECAM-1 gene.
Hypoxia also activates the transcription factor NF-kB [16], which promotes the production of several molecules. NFkB increased TNFa [17], which is an inflammatory cytokine, as well as ICAM-1 [18], which is an adhesion molecule also involved in inflammation. The chemokines MCP1 and MIP1B, as well as the cytokine IL1B, are also activated by NFkB [19] and play an important role in the migration of monocytes to fight infection.
Hypoxia inhibits the transcription of the MARCH5 gene, which is known to activate the PI3k/Akt pathway and increase the production of eNOS [20]. Hypoxia also increases the production of the protein kinases p38 and p62 [21], which promote HIF1a and VEGF [22] while attenuating eNOS production [23].
[1] L. Flamant, S. Toffoli, M. Raes, C. Michiels, Hypoxia regulates inflammatory gene expression in endothelial cells (https://pubmed.ncbi.nlm.nih.gov/19101540/)
[2] D.J. Manalo, A. Rowan, T. Lavoie, L. Natarajan, B.D. Kelly, S.Q. Ye, J.G.N. Garcia, G.L. Semenza, Transcriptional regulation of vascular endothelial cell responses to hypoxia by HIF-1 (https://pubmed.ncbi.nlm.nih.gov/15374877/)
[3] A. Mammoto, K. Hendee, M. Muyleart, T. Mammoto, Endothelial Twist1-PDGFB signaling mediates hypoxia-induced proliferation and migration of αSMA-positive cells (https://pubmed.ncbi.nlm.nih.gov/32371931/)
[4] B. Zhang, W. Niu, H.Y. Dong, M.L. Liu, Y. Luo, Z.C. Li, Hypoxia induces endothelial‑mesenchymal transition in pulmonary vascular remodeling (https://pubmed.ncbi.nlm.nih.gov/29568878/)
[5] https://www.uniprot.org/uniprot/P09619
[6] A.P. Laddha, Y.A. Kulkarni, VEGF and FGF-2: Promising targets for the treatment of respiratory disorders (https://pubmed.ncbi.nlm.nih.gov/31421589/)
[7] L. Li, M. Xu, X. Li, C. Lv, X. Zhang, H. Yu, M. Zhang, Y. Fu, H. Meng, J. Zhou, Platelet-derived growth factor-B (PDGF-B) induced by hypoxia promotes the survival of pulmonary arterial endothelial cells through the PI3K/Akt/Stat3 pathway (https://pubmed.ncbi.nlm.nih.gov/25613241/)
[8] C.S. Abhinand, R. Raju, S.J. Soumya, P.S. Arya, P.R. Sudhakaran, VEGF-A/VEGFR2 signaling network in endothelial cells relevant to angiogenesis (https://pubmed.ncbi.nlm.nih.gov/27619687/)
[9] J. Cheng, H.L. Yang, C.J. Gu, Y.K. Liu, J. Shao, R. Zhu, Y.Y. He, X.Y. Zhu, M.Q. Li, Melatonin restricts the viability and angiogenesis of vascular endothelial cells by suppressing HIF-1α/ROS/VEGF (https://pubmed.ncbi.nlm.nih.gov/30569127/)
[10] J.M. Hardwick, L. Soane, Multiple Functions of BCL-2 Family Proteins (https://pubmed.ncbi.nlm.nih.gov/23378584/)
[11] U. Forstermann, T. Munzel, Endothelial nitric oxide synthase in vascular disease: from marvel to menace (https://pubmed.ncbi.nlm.nih.gov/16585403/)
[12] P. Gao, N. Niu, T. Wei, H. Tozawa, X. Chen, C. Zhang, J. Zhang, Y. Wada, C.M. Kapron, J. Liu, The roles of signal transducer and activator of transcription factor 3 in tumor angiogenesis (https://pubmed.ncbi.nlm.nih.gov/28978186/)
[13] K.S. Kim, V. Rajagopal, C. Gonsalves, C. Johnson, V.K. Kalra, A Novel Role of Hypoxia-Inducible Factor in Cobalt Chloride- and Hypoxia-Mediated Expression of IL-8 Chemokine in Human Endothelial Cells (https://pubmed.ncbi.nlm.nih.gov/17082639/)
[14] M. Bickel, The role of interleukin-8 in inflammation and mechanisms of regulation (https://pubmed.ncbi.nlm.nih.gov/8315568/)
[15] J. Wang, R. Zohar, C.A. McCulloch, Multiple roles of alpha-smooth muscle actin in mechanotransduction (https://pubmed.ncbi.nlm.nih.gov/16325810/)
[16] N. Hashimoto, K. Ikuma, Y. Konno, M. Hirose, H. Tadokoro, H. Hasegawa, Y. Kobayashi, H. Takano, DPP-4 inhibition protects human umbilical vein endothelial cells from hypoxia-induced vascular barrier impairment (https://pubmed.ncbi.nlm.nih.gov/28923269/)
[17] N.S. Trede, A.V. Tsytsykova, T. Chatila, A.E. Goldfeld, R.S. Geha, Transcriptional activation of the human TNF-alpha promoter by superantigen in human monocytic cells: role of NF-kappa B (https://pubmed.ncbi.nlm.nih.gov/7608567/)
[18] H. Patel, N. Zaghloul, K. Lin, S.F. Liu, E.J. Miller, M. Ahmed, Hypoxia-induced activation of specific members of the NF-kB family and its relevance to pulmonary vascular remodeling (https://pubmed.ncbi.nlm.nih.gov/28987523/)
[19] S. Kochumon, A. Wilson, B. Chandy, S. Shenouda, J. Tuomilehto, S. Sindhu, R. Ahmad, Palmitate Activates CCL4 Expression in Human Monocytic Cells via TLR4/MyD88 Dependent Activation of NF-κB/MAPK/ PI3K Signaling Systems (https://pubmed.ncbi.nlm.nih.gov/29669317/)
[20] W. Lei, J. Li, C. Li, L. Chen, F. Huang, D. Xiao, J. Zhang, J. Zhao, G. Li, T. Qu, H. Zhou, Y. Liao, M. Chen, MARCH5 restores endothelial cell function against ischaemic/hypoxia injury via Akt/eNOS pathway (https://pubmed.ncbi.nlm.nih.gov/33611830/)
[21] N. Jin, N. Hatton, D.R. Swartz, X.I. Xia, M.A. Harrington, S.H. Larsen, R.A. Rhoades, Hypoxia activates jun-N-terminal kinase, extracellular signal-regulated protein kinase, and p38 kinase in pulmonary arteries (https://pubmed.ncbi.nlm.nih.gov/11062137/)
[22] M.C.A. Duyndam, S.T.M. Hulscher, E. van der Wall, H.M. Pinedo, E. Boven, Evidence for a role of p38 kinase in hypoxia-inducible factor 1-independent induction of vascular endothelial growth factor expression by sodium arsenite (https://pubmed.ncbi.nlm.nih.gov/12482858/)
[23] R.P. Weerackody, D.J. Welsh, R.M. Wadsworth, A.J. Peacock, Inhibition of p38 MAPK reverses hypoxia-induced pulmonary artery endothelial dysfunction (https://pubmed.ncbi.nlm.nih.gov/19201999/)
Meeting Notes:
- Henrique had some comments about our combined model:
- We should have PI3k –> STAT3 –> VEGF instead of PI3k –> VEGF
- There is probably an intermediate in the connection from VEGFR2 –> BMP2/4
- There is also likely an intermediate in BMP2 –> ROS/eNOS
- BMP2 and BMP9 are shown to have opposite effects in the literature, so we should try to see if the code shows this
- Instead of having BMP9 –| Akt directly, BMP9 probably activates PTEN, which inhibits Akt (but we should do our own research)
- Some molecules (like Ras) are always on if they’re not being inhibited
- Our code suggests that we might have attractors in our network (when you stay in a cycle in the network and the state doesn’t change)
Homework:
Look into the suggestions Henrique had and also look at the attractors.
07/01/2022 - Attractors
Background:
This week, we made a list of potential attractors by looking at which nodes always stayed on when we ran the code.
When we ran the code with hypoxia on, the following were the attractors:
HIF1a, IL8, VEGF, TNFa, IL1b_MIP1b, CD31, Twist1, aSMA, Col, PDGFB, Bcl2, MARCH5,
eNOS, ROS, NFkB, ICAM1, Ras, BMP9, SMAD1, PDGFR, BMPR_ALK, Raf, MEK1_2,
SMAD6_7, CXCL8, PTEN, MCP1
When we ran the code with shear stress on, the following were the attractors:
HIF1a, IL8, VEGF, TNFa, IL1b_MIP1b, CD31, Twist1, aSMA, Col, PDGFB, Bcl2, MARCH5,
eNOS, ROS, NFkB, ICAM1, Ras, BMP2_4, BMP9, SMAD1, PDGFR, BMPR_ALK, Raf=31,
MEK1_2, SMAD6_7, CXCL8, PTEN
I also read some papers and updated some connections:
https://pubmed.ncbi.nlm.nih.gov/12447987/
- ERK & Hypoxia together activate HIF1a
- If ERK is inhibited, hypoxia doesn’t activate HIF1a. Hypoxia needs ERK to activate HIF1a
https://pubmed.ncbi.nlm.nih.gov/24614941/
SMAD1 inhibits ERK
https://pubmed.ncbi.nlm.nih.gov/15369676/
ROS → HIF1a even in normoxia
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC33561/
PTEN –| Akt
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1986742/
- PTEN –| Akt (confirmed)
https://www.researchgate.net/figure/BMP9-and-BMP10-induce-vascular-quiescence-by-various-mechanisms-Through-ALK1_fig2_338439506
BMP9 → PTEN
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7390625/
BMP9 –| MCP1 (CCL2)
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2708876/
BMP9 → CXCL8 (ALK1 necessary)
Meeting Notes:
- Henrique said we should try running the code with Hypoxia and Shear always on, and with BMP9 always off, since that is how most experiments in vitro are run
- Since BMP2 and BMP9 have opposite effects, it is possible that ALK1 –> SMAD and ALK2 –> PTEN
- SMAD may activate Akt
Homework:
We should try to find a new list of attractors with the new conditions and read about some of the connections he mentioned.
07/08/2022 - Attractors
Background:
This week, I reran the code with hypoxia/shear always on and BMP9 off, and this is what I got:
Run with hypoxia on:
Run with shear on:
This is what the weird graph looked like:
Meeting Notes:
- Melody and Henrique said that the fluctuations were interesting. We ran it multiple times with ensemble size = 1, and it showed that individually, it sometimes fluctuated and sometimes didn’t. Every time, the graph is a little different.
Homework:
Our homework is to keep looking for intermediates between the BMPs and other genes/molecules.
07/15/2022 - Intermediates
Background:
This week, I tried to find out how BMP9 leads to PTEN. BMP9 is an extracellular signalling molecule, so it can’t go directly to PTEN and must go through a receptor.
https://pubmed.ncbi.nlm.nih.gov/29449337/
- BMP9/ALK1 –> PTEN
https://pubmed.ncbi.nlm.nih.gov/16418168/
- BMP9 –> p38
- p38 –> ATF2 –> PTEN
So, based on these papers, the pathway looks like this:
BMP9 → ALK1 → p38 → ATF2 → PTEN
Meeting Notes:
- Henrique said that this pathway made sense. Since BMP9 and BMP2 usually have opposite effects, it wouldn’t make sense for BMP9 –> PTEN to go through SMAD because the BMPs all go through SMAD. It made more sense to have a different transcription factor that only BMP9 could activate.
Homework:
Our homework was to look for more intermediates and also see if our input/output data matches the code.
07/22/2022 - Validating
Background:
This week, I simulated the experiments run in the papers through my code to see if the results matched. I added two columns to my input/output spreadsheet to track this:
Meeting Notes:
- We discussed what increase/decrease means in the context of a boolean model. Since the only two states in our model are on and off, it’s hard to define increase/decrease. Increase can still make sense if something is activated, but something can’t decrease if it’s already off and doesn’t turn on. We have to think about alternatives, or we could just say that our model “doesn’t contradict” the data
Homework:
Our homework is to combine the written parts (intro and description).
07/29/2022 - Updating Introduction
Background:
This week, we updated our introduction by combining the separate drafts that we already had. We tried to make it more concise and integrate both hypoxia and shear stress, though we did not put in our references yet.
Meeting Notes:
- Melody found new datasets that she and Henrique will look at to see if we can use them to validate our network
- The datasets she found had pulmonary microvascular ECs exposed to hypoxia, and data was recorded at multiple time points instead of just one, which gives a better picture
- We discussed how we could list limitations of our boolean model in a section of the paper (like the fact that it is hard to model “increase” and “decrease”)
- One possibility for making the input/output validation still work is to have a random 10% of the nodes initially on instead of only turning on a single node at the beginning. This may better simulate random initial conditions than just relying on the asynchronous nature pf the model for randomness.
Homework:
Our homework is to combine our separate descriptions of the network into one cohesive draft. We should also think about the other limitations of our model that we could list. I also have to update my input/output sheet: instead of just “match” or “no match”, I should put the value at which the network stabilized. I need to do this three times: once with the experimental conditions, once with a random 10% of the nodes initially on, and once with both of the previous conditions (so we can compare).
08/05/2022 - Methods Section
Background:
This week, we worked on the new input/output data we needed. Here is some of the table:
We also combined both of our model descriptions to make it about both hypoxia and shear stress.
Meeting Notes:
- The methods section should discuss how we built our model, not the model itself
- We could try using Overleaf/Latex so the citations will be easier
- There should be a section about limitations in our methods section
- We could read other papers that used boolean models for cell signaling to see how they did their papers
- There should be a table with all of our equations and citations in it, and another table with the abbreviations we used.
Homework:
Fix our methods section, read similar papers, and make the tables.
08/12/2022 - Editing Methods Section
Background:
This week, we fixed our methods section. We first read some papers that also used boolean models for cell signaling to see how their methods sections were structured.
Some of the papers we read were:
Weinstein: https://www.frontiersin.org/articles/10.3389/fphys.2017.00960/full
Wang: https://royalsocietypublishing.org/doi/10.1098/rsta.2019.0338
Here is some of our new methods section (the whole thing is really long):
To create our model, we first focused on the direct downstream effects of
hypoxia and shear stress on ECs and the molecules that are involved in caus-
ing the changes that were observed. We characterized the signaling pathways
involving key transcription factors such as HIF1a and NFkB and identified
the genes that were upregulated and downregulated in response. We then ana-
lyzed other connections between the genes and molecules we already had in
order to gain a more complete understanding of how the whole system inter-
acts.
I also added citations to the introduction, which was easier now in LaTeX. I tried making the tables, but I was having trouble with the formatting in Overleaf.
Meeting Notes:
- We talked about data validation and the data Melody found.
- It was from pulmonary endothelial cells, and they were HMVECs (which is what we want), so that was good. However, the adjusted p-values were very high because there were only 3 samples for each gene. When all the timepoints were lumped together, there were 3 genes that were differentially expressed. When pulmonary and cardiac cells were also lumped together, there were 24 genes that were differentially expressed, but putting different cell types together may not be good.
- Another dataset that Melody found may be better, and it has a lot of genes that were differentially expressed.
- Many of the genes that are shown in our network to be upregulated are not in the lists of genes in the data. Henrique suggested that this may be due to Type II Error (they were just excluded because there’s so much randomness in RNA sequencing), so it may be better to just use the gene lists to look for transcription factors using ChEA3.
Homework:
Our homework is to make the tables after Melody helps us format them, look for more new papers to use for input/output validation, and play around with the datasets.
08/19/2022 - Table of Equations
Background:
This week, I spent time on the table of equations for the paper. The citations were taking me a long time, so I still have some of those left. Below are the first few rows of the table:
I also separated STAT1 and STAT3 (which we had together before) because I read that they actually had opposite effects.
Meeting Notes:
- I asked if I should leave the rows that just had “Hypoxia = Hypoxia” or “BMP9 = BMP9”, and Melody told me to keep them and just put “fixed” in the reference column
- I asked about citing reviews if they didn’t reference an experiment in the review – Henrique and Melody said that it was fine as long as I tried to look
- Some notes about the Miro
- GIT isn’t a receptor – it’s a gene that comes from the receptor GPCR (G-protein coupled receptor)
- We should keep either p38 or p62 in the equations and just make a note at the bottom (since the equations should be simple). The Ca2+ ion could also just be written as Ca to simplify.
- There is a linear pathway with Ras to Raf to MEK to ERK. We had a connection with Ras to MEK, but Raf is the intermediate, so we can get rid of that.
- “Regulates” is vague, so we should specify that BCL2 inhibits apoptosis
- We should combine the effects (we have apoptosis in multiple effects boxes, so instead we should make one box and have all the pathways that relate to it leading there). Melody found lots of papers on how hypoxia is related to apoptosis that we can look at.
- GIT1 doesn’t lead anywhere anymore, so Melody found a good paper we could look at.
- SMAD1 is a transcription factor (which is what we have), but SMAD6/7 are genes (we had them as transcription factors as well). They are actually genes that inhibit the other SMADs.
- Melody also found the official gene names for all of our genes, made a list, and put it into ChEA3.
- HIF1a, NFkB1/2, and Twist1 were in the top 100 and in our model
- Twist2, ATF3, and SMAD4 were also in the top 100. They’re not in our model, but they’re similar to transcription factors we do have.
- SMAD1, ATF2, and STAT1/3 were not in the top 100, but they were in our model, so we might need to look into those.
Homework:
Finish the table and make the changes to the model.
08/26/2022 - Finishing Citations
Background:
This week, I finished adding citations for all the equations/connections that I had/could find the papers for. I also combined the effects: instead of having each molecule go to some effects, I combined them and had 3 main effects (angiogenesis, apoptosis, and vasodilation) with different molecules either promoting or inhibiting them. I read two of Melody’s papers on apoptosis and found that p38 is pro-apoptotic while ERK is anti-apoptotic, so I added those.
Meeting Notes:
- Henrique and Melody said it was okay to cite this paper (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC316444/) for SMAD6 inhibiting SMAD1 even though it is not specific to endothelial cells because SMAD6 –| SMAD1 is a well-established fact
- Another effect we could add is increased permeability (the cell-cell junctions are open) – eNOS and ROS increase permeability while PECAM1 inhibits it (instead stabilizes the barrier)
- We can look more into the main effects we had and make sure we have a complete picture so we can talk about that in our methods section
- We had some arrows going directly from VEGF or PDGFB to other genes instead of going through VEGFR and PDGFR, so we fixed that
- STAT1 has no molecules coming from it so we could add some (pro-inflammatory cytokines)
- Even though Bax increases apoptosis and BCL-X1 inhibits it, we don’t need to separate the BCL2 in the network; instead, we can just find which one is activated by hypoxia
Homework:
My homework is to find papers for the rest of the table citations, find molecules coming from STAT1, and looking more into hypoxia and apoptosis.
09/02/2022 - STAT1 & Apoptosis
Background:
This week, I completed all the citations in the table. Since our network is still changing, I might need to change some of the equations in the future though.
I also found connections for STAT1, which basically has the opposite effects as STAT3.
https://www.ahajournals.org/doi/10.1161/atvbaha.109.192542
- STAT1 inhibits HIF1a
https://pubmed.ncbi.nlm.nih.gov/16585190/
- STAT1 inhibits VEGF & angiogenesis
I read some more of the apoptosis papers and found BCL2 and Bax having opposite effects, but I wasn’t sure what was turned on during hypoxia. I also couldn’t figure out whether hypoxia led to apoptosis or of other antiapoptotic molecules prevented apoptosis during hypoxia.
Meeting Notes:
- Henrique said to double-check the connection that COX2 leads to angiogenesis; he said it could lead to vasodilation
- Melody found a paper that said COX2 came from VEGF (which is increased by hypoxia), so I will read that paper and have COX2 come from VEGF instead of directly from hypoxia.
- Henrique has a paper about BMP9 and what it activates/inhibits: https://www.ahajournals.org/doi/10.1161/ATVBAHA.118.310760
- I had some questions about the citations that Melody and Henrique answered
- I couldn’t find any papers that had EGFR –> Ras/Raf but I found the following paper that mentioned the “EGFR/Ras-signalling pathway.”
https://www.ahajournals.org/doi/10.1161/ATVBAHA.118.310760- They said to look up EGFR and Ras on Kegg to make sure there weren’t any interesting connections to other molecules and that it fine to cite the paper I found.
- I couldn’t find a paper showing that low shear stress inhibits COX2, so Melody found some papers about COX2 that might help
- The paper I found for SMAD –| ERK used weird stem cells from zebrafish. Melody said to check if the stem cells were converted into endothelial cells (in which case the paper is okay to use).
- I couldn’t find any papers that had EGFR –> Ras/Raf but I found the following paper that mentioned the “EGFR/Ras-signalling pathway.”
Homework:
My homework is to read about COX and its effects, read the BMP9 paper, check the SMAD/ERK paper, look at EGFR/Ras signaling, and find papers about vasoconstriction/vasodilation during hypoxia and shear stress.
09/09/2022 - Vasoconstriction
Background:
Here is what I did this week:
COX2 Papers
- COX2_96 (HUVECs)
- Inflammation, proliferation
- Hypoxia → NFkB → COX2
- COX2_06 (HUVECs)
- VEGF & p38 → COX2 (both needed)
- Angiogenesis
- VEGF → p38
- COX2_03 (HUVECs)
- Angiogenesis
- VEGF → COX2
- IL1B → COX2
- https://www.ahajournals.org/doi/full/10.1161/hc4901.101350
- COX2 does not affect vasodilation
- Much more complete now but still nothing about low shear stress
**Everything comes from VEGFR in Miro → need to replace all VEGF with VEGFR in the table? (e.g. ROS = VEGFR instead of ROS = VEGF)
Henrique’s Paper: https://www.ahajournals.org/doi/10.1161/ATVBAHA.118.310760
- ALK1 → BMP9
- BMP9 → PTEN –| PI3k
- ALK1 → SMAD1,5,8
https://www.kegg.jp/pathway/map04014
- Couldn’t find intermediate between EGFR and Ras (RasGFR) so kept previous citation referencing the EGFR/Ras pathway
https://www.nature.com/articles/ncomms4431
- Seems to use endothelial cells for SMAD1 –| ERK
- “Transverse sectioning post WISH showed that the expression of erk1 and erk2 in the endothelial cells (red arrows in the black square) was increased in embryos injected with smad1 MO or smad5 MO”
Vasoconstriction papers for hypoxia:
https://www.sciencedirect.com/science/article/pii/S2211383512001669
- NO is a vasodilator confirmed (this is a review though)
- Hypoxia → vasoconstriction to raise arterial blood pressure
- COX2 → ROS (“other enzymes that can produce ROS”)
- ROS → vasoconstriction
- Says that the involvement of ET1 is doubtful but other papers say it leads to vasoconstriction
https://pubmed.ncbi.nlm.nih.gov/2545192/
- ET1 → vasoconstriction
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5301302/
- NO → vasodilation
- Ca2+ → vasoconstriction
- ROS can increase or decrease during hypoxia
- In pulmonary vascular smooth muscle cells, ROS increases and causes calcium to increase and cause vasoconstriction
ROS is confusing!!!
https://pubmed.ncbi.nlm.nih.gov/18339777/
- States that some say that hypoxia increases ROS, other say decreases ROS
- ROS → decreases during hypoxia & normally causes vasodilation
- But the other paper said vasoconstriction??? And that ROS increases during hypoxia???
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5301302/
- “This controversy regarding ROS during hypoxia might be related to the duration of hypoxia. ROS production by human PASMCs attenuates initially when exposed to brief period of hypoxia [65] but increases subsequently”
- NO (eNOS) decreases during hypoxia, inhibition of NO augments vasoconstriction
Vasoconstriction in low shear stress:
https://pubmed.ncbi.nlm.nih.gov/12577139/
- Low shear = turbulent flow? Maybe use the alternate term to find more papers?
- Low shear promotes vasoconstriction
- No access beyond abstract
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4220438/
- ET1 → vasoconstriction
Can’t find more shear papers but probably similar to hypoxia?
Remaining Citation Issues:
- Calcium shear citation uses fluid shear stress not LSS
- COX2 shear stress citation missing
- COX2: keep both hypoxia and NFkB or just NFkB?
- GIT1 citation doesn’t work
Meeting Notes:
- During the meeting, I talked about what I did this week and asked my questions
- Henrique said that I was correct that COX2 doesn’t affect vasodilation (he said that the paper he was thinking of was about neurons)
- I should change VEGF to VEGFR in all the equations in the table
- I had one paper saying ALK1 –| SMAD1 while another said that ALK1 –> SMAD1. Henrique and Melody said that ALK1 should probably increase SMAD1, so they both sent me papers I can look at and I should probably end up changing it to increase.
- Henrique confirmed that NO is the same as eNOS (it’s a product of eNOS)
- They both agreed that the amount of ROS during hypoxia depended on the duration of the hypoxia as stated in one of the papers. We still have not decided if we’re going to focus on the immediate effects of hypoxia or look at continuous hypoxia and so we don’t know yet which one to keep.
Homework:
My homework this week is to write about what I found regarding apoptosis and vasodilation for our methods section so we can focus the story of the paper and have the pathways we looked at that dealt with each overall effect of hypoxia and low shear.
09/16/2022 - Methods Section
Background:
This week, I edited the methods section to frame it around the four effects we found: angiogenesis, permeability, vasoconstriction, and apoptosis.
Meeting Notes:
- Melody and Henrique said to talk less about the biology in the methods section and have fewer citations, since the methods should be more about what we actually did.
- We should also have a section in the methods where we talk about how we created our Boolean equations and what all the symbols mean
- We should also add a description column to our table of equations like the ones in some other signalling papers
Homework:
Our homework is to write the introduction and methods sections of the paper
09/23/2022 - Revised Methods
Background:
This week, I revised the methods section to focus more on what we actually did, like Melody wanted. I also came up with descriptions/explanations for the equations in the table, but I couldn’t put them into Overleaf because the formatting was messed up. Here are some examples:
- HIF1 is activated by hypoxia together with p38, ERK, or ROS, or by either hypoxia or Akt independently.
- IL8 is activated by the HIF1 or NFkB transcription factors, or by the ALK1/BMP9 pathway.
Meeting Notes:
- We talked about revisions on the writing portion
- Henrique shared a website that lets you generate LaTeX tables more easily so I can add the descriptions into Overleaf: https://tablesgenerator.com/
- We talked about updating the code
- For the genes that are differentially regulated under hypoxia, we should start them opposite to where they end up. Instead of starting all of the nodes (except hypoxia or shear) at 0, we should start certain nodes at 1 (the nodes that decrease or are inhibited in response to hypoxia/shear — get this from the input/output spreadsheet). This way, the nodes that increase under hypoxia start at 0 and go to 1, and the nodes that decrease start at 1 and go to 0 (instead of starting and staying at 0).
- Henrique and Melody showed me a way to turn on only certain nodes in R so I can do this
Homework:
This week, my homework is to add the descriptions to the Table of Equations in Overleaf, update the code with the current equations, and change the initial values of the nodes that are supposed to decrease.
10/07/2022 - Updating Code
Background:
This week, I tried to add the descriptions to the table using the website Henrique gave me, but it still wasn’t formatting it correctly in Overleaf.
I also updated the code with what was in the table/Miro. The papers we had said that CD31 (PECAM1), MARCH5, and eNOS were downregulated in response to hypoxia, so I initialized those values as 1 in the code. None of the molecules we had were directly downregulated by shear, so all of those started at 0. This was what one of our graphs looked like:
I also added some descriptions of the code to the methods section.
Meeting Notes:
- Melody and Henrique clarified that only molecules that were explicitly stated in papers to be downregulated by hypoxia/shear should be initialized to 1. If it is decreased in our model but not mentioned in papers, we start it at 0 like everything else.
- Henrique showed me how the website could wrap text to help the table look better in Overleaf, and Melody also said she would try to fix it.
- Starting a molecule at 0 vs 1 still keeps the same meaning in the graph. For example, when ERK started at 0, it created the following graph (under hypoxia):
When ERK started at 1, the graph changed to look like this:
However, these are basically still the same graph, it’s just that the first few timepoints are different because of the different initial conditions. There is still a peak at 0.4/0.5 for both (burst of upregulation) and they both go down to 0 for the rest of the timesteps.
Homework:
Our homework is to run the input/output simulations again with the updated code, think about what questions we could ask with/about our model, try to fix the table, and make the Miro look more organized. Also, we should include quantitative information about our network (number of nodes, how many transcription factors and what they are, etc.) in the methods.
10/14/2022 - Input/Output Validation
Background:
This week, I ran the code with the experimental conditions from the input/output papers and recorded whether or not the outputs matched. These are some of the rows (the blue shading means the outputs don’t match).
I also added the quantitative information to the methods section, such as the number of each type of node.
Meeting Notes:
- Melody was able to fix the table formatting
- There was one paper that said that knockdown of HIF1a decreases VEGF, but when I ran the code with HIF1a knocked down, VEGF was still on (at 1). We looked through the paper and one of the graphs showed that under hypoxia, knockdown of HIF1a causes VEGF to increase LESS. So, VEGF still increases to above-normal levels even with HIF1a knocked down; it’s just a smaller increase than with HIF1a active.
Homework:
This week, I will change/edit some of our papers so they are all about low shear stress, finish running code simulations, finish adding descriptions to the table, and investigate why some of the outputs don’t match by rereading the papers.
10/21/2022 - Finished Table
Background:
This week, I finished adding descriptions to the table and reread the following paper that was in our input/output sheet: https://pubmed.ncbi.nlm.nih.gov/25613241/
The paper first showed that hypoxia increases PDGFB, then showed that PDGFB increases STAT3 and Akt. So the input/output should be PDGFB –> STAT3, Akt and not Hypoxia –> STAT3, Akt. This may be the reason the result of the code didn’t match.
Meeting Notes:
- Henrique said to double-check that MARCH5 and CXCR4 are always on except when Hypoxia and ALK1 are on, respectively.
- There was a duplicate citation in our bibliography
- We changed the descriptions for “hypoxia = hypoxia” and “shear = shear” from “fixed” to “fixed by testing environment”
- Melody said that after we finished validating our model, we would try turning receptors on and off to see what the effects are so we can maybe say something about our model.
Homework:
The papers in the table citations don’t match the “old papers” sheet in the input/output spreadsheet. We need to:
- delete the rows with molecules that are not in our network
- move the rows with papers we didn’t end up using to build to model to the “new papers” sheet
- add in all the papers we have cited in the Overleaf table that are not already in the input/output sheet
One we’re done with this, we should also rerun the model with the updated inputs and outputs.
10/28/2022 - Editing Input/Output
Background:
This week, we went through our input/output sheet and made sure the citations in Overleaf matched the sheet and fixed any inconsistencies.
Meeting Notes:
- We discussed another Student Research Symposium that we will present at
- We talked about the process of validating with the code
Homework:
Finish updating the input/output table with both old and new papers, and run the model simulations.
11/04/2022 - Validation
Background:
This week, we finished updating all the cited papers. I also edited the code a little and ran some new simulations. Most of the simulation results matched the input/output.
For the Kim2006 paper, the model showed increased IL8 for Hypoxia + Akt inhibitor, but in the paper IL8 decreased. After rereading the paper, I found that IL8 still increased relative to normoxia — it was just less of an increase.
Meeting Notes:
- Henrique said to try to find papers that change (turn on/off) the receptors in our model and see what effects that has on our phenotypes (angiogenesis, apoptosis, etc.)
- Melody said to have a “story” about our model for the next meeting (something interesting we found about it)
Homework:
We should finish running the model simulations, find new papers for the receptors/phenotypes, and have something about the model to share.
11/11/2022 - Chaos!
Background:
This week, Isabella and I ran our model with all the different inputs for the papers and checked whether or not the outputs matched. The majority of them did match, though some didn’t.
For one of our graphs (plotting ERK with the inhibition of MEK), we got this:
20 iterations:
100 iterations:
500 iterations:
Meeting Notes:
- Melody and Henrique weren’t sure why the graph came out like this
- It seemed like chaos (which is when a small change in the input causes a big change in the output)
- Melody showed us a graph of the discrete logistic model (x = rx(1-x)) and its bifurcations as an example of this, and how a small change in r could completely change the result
- This is how Henrique’s plot of the logistic function changed with slightly different values of r: (first two repeat – bifurcations; last one is chaos)
- Henrique told us about the Lorenz equations and how a tiny change in the rounding could change the weather prediction
- Melody showed us a graph of the discrete logistic model (x = rx(1-x)) and its bifurcations as an example of this, and how a small change in r could completely change the result
- The model is stochastic (random) because a random node is updated at each timestep and there are many possible states
- (As opposed to deterministic, where an initial state only has one end state)
- The graph was slightly different each time the function was run, and it didn’t seem to be repeating
- It seemed like chaos (which is when a small change in the input causes a big change in the output)
Homework:
This week, our homework is to check the results that weren’t consistent with the papers. For example, we should check if some of the inputs happened under hypoxia and we just forgot to add that in our simulation.
11/18/2022 - Reviewing Inconsistencies
Background:
This week, we went through the results that didn’t match what the experiments said to try to figure out if our model needs to be edited. Some changes were minor, but there were two questions I came across:
- One paper that we used in our model said that hypoxia activates ROS, but another paper said that hypoxia activates NFkB, which inhibits ROS. Therefore, when we ran our model with hypoxia as the only input, ROS decreased (since hypoxia was activating NFkB). This contradicted the first paper, which said hypoxia –> ROS
- One paper said that ALK1 inhibition increases VEGFR2. However, in our model, we had VEGFR2 being activated by VEGF and not ALK1. So, when the model was run with ALK1 inhibition as the only input, VEGFR2 was not activated (the presence of VEGF would have also been required for our model to activate VEGFR2).
Meeting Notes:
- For the first question:
- Melody said that both papers may be true but the boolean model may just not be able to represent this. For one thing, the two experiments were run under different conditions by different people, so the first experiment may have had lower NFkB levels, allowing ROS to increase during hypoxia, or there might have just been other differing experimental conditions causing this. Also, it may be that hypoxia does increase ROS, but less so in the presence of NFkB. However, the boolean model can only show on or off, not relative expression levels (increase/decrease), so the model may just not be able to show the results.
- For the second question:
- Henrique said that many papers measured the expression of a receptor, while we were concerned with the activation of the receptor. In the paper I found, it may be that ALK1 inhibition results in more VEGFR2 receptors being made/produced. However, in our model, we were measuring activation (whether something binds to the receptor and makes it active/on), which is different from “is the receptor present at high levels?”. Therefore, our model and the paper don’t agree, but they don’t disagree either (they’re just measuring two different things), so it’s fine.
- There was a bug in our code that was making the edited version of the model output values greater than 1.
Homework:
Our homework is to finish checking the inconsistencies in the paper results.
12/02/2022 - Abstract
Background:
This week, we wrote our abstract for the Student Research Symposium and titled it: A Boolean Network Simulating Low Shear Stress and Hypoxic Cell Signaling Pathways in Pulmonary Endothelial Cells
Meeting Notes:
- Henrique went over what we should include in our presentation:
- Introduce our topic and project
- Picture of the Boolean network in Miro
- Heatmap showing the states of all the different nodes
- One of our individual graphs that is “interesting”
- Comparison with the literature
Homework:
This week, we should create the presentation for the research symposium.
